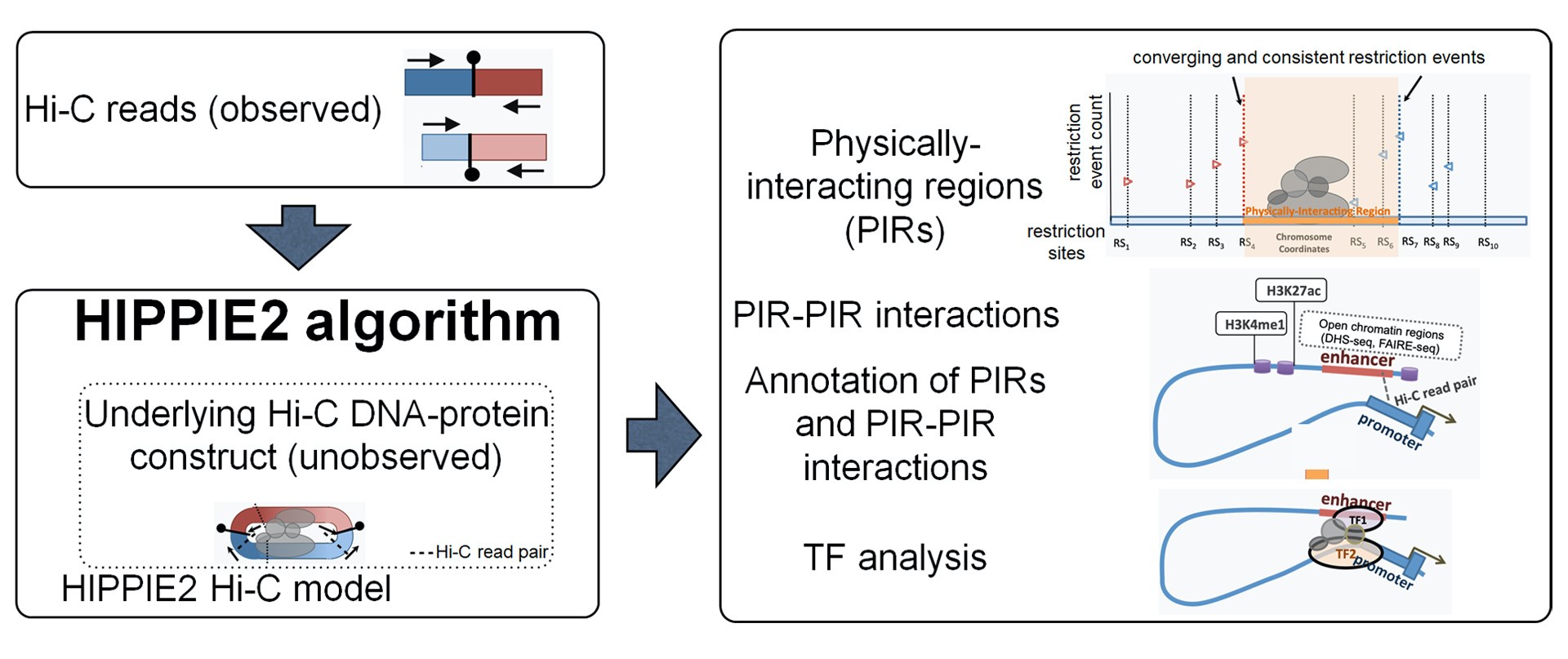
PNGC is pleased to announce the release of “HIPPIE2: a method for fine-scale identification of physically interacting chromatin regions”. Because most regulatory chromatin interactions are mediated by numerous transcription factors (TFs) and involve physically interacting regions (PIRs), HIPPIE2 was developed to map these elements and interactions at a much finer scale (sub-1Kb resolution).
HIPPIE2 is aimed at improving analysis precision and resolution of high-throughput chromosome conformation (Hi-C) experiments to identify precise loci of DNA physically interacting regions (PIRs).
Unlike genome binning approaches with fixed-size genomic bins that potentially contain multiple interaction sites, HIPPIE2 introduces a dynamic, non-binning algorithm that infers the physical locations of PIRs using multiple sources of information including restriction event distribution, inferred relative location of protein-bound interaction sites, mapped read locations and orientations. HIPPIE2 thus provides a more accurate, high-resolution map of chromatin and regulatory interactions and mediating TFs.
HIPPIE2 source code is available here: https://bitbucket.org/wanglab-upenn/hippie2.
HIPPIE2 has been published in NAR Genomics and Bioinformatics this March. The full manuscript can be found here: https://doi.org/10.1093/nargab/lqaa022.
PNGC congratulates the authors, Pavel P. Kuksa, Alexandre Amlie-Wolf, Yih-Chii Hwang, Otto Valladares, Brian D. Gregory, and Li-San Wang, for their hard work!