
Genome sequencing results can be used to train PRS models but suffer from computation and privacy limitations. A new web portal from Drs. Jin Jin and Bingxin Zhao uses GWAS summary statistics and novel pseudo-training methods to make the latest generation of PRS models more accessible.
Your DNA impacts your risk for many diseases, such as Alzheimer’s disease, cancer, and schizophrenia. This risk-level can be useful for life-planning and medical treatment decisions. However, disease prediction isn’t as simple as testing for a “cancer gene” or an “Alzheimer’s gene”. Accurate prediction of disease requires sophisticated models that can account for sex, human ancestries, and complex inheritance patterns that simple gene tests would miss.
Polygenic risk scores (PRS) add up thousands or even millions of variations in the DNA and combine these into a prediction of how likely it is that an individual has a specific trait or disease. By comparing the genetics of people with and without a disease, the variants with the greatest impacts on disease risk can be determined.
PRS models are often built to specialize on a single disease or set of diseases. However, the design of a PRS model (i.e., how many variants should be used, what kind of statistical modeling should be performed, etc.) built to predict one disease may be useful to researchers with data describing another. However, access to this genetic data is often highly restricted. One solution would be for researchers to train cutting edge PRS models on their new data, but such models can require computational resources and expertise. An alternative approach is to use summary statistics instead.
At the University of Pennsylvania, Dr. Jin Jin, Assistant Professor of Biostatistics, and Dr. Bingxin Zhao, Assistant Professor in the Wharton Department of Statistics and Data Science, are working to help researchers bring their data to cutting edge PRS models without requiring individual-level genetic data to be shared. Their team has developed PennPRS (pennprs.org), a new cloud computing platform that can perform PRS training online using only summary statistics. This will enable researchers to create new PRS models, making prediction possible for new traits, diseases, and populations. Such models only made possible by alleviating cost, expertise, and privacy barriers.
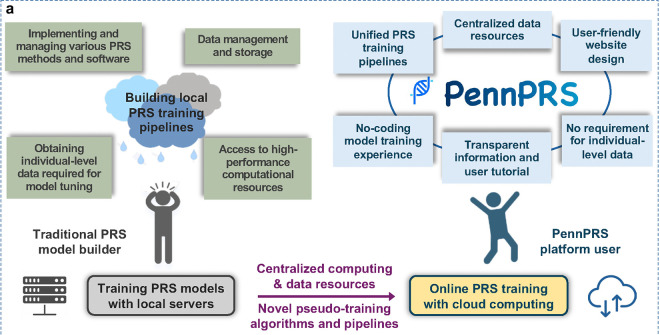
Fig. 1: The Challenges of Traditional PRS Model Training and the Promise of PennPRS Cloud Computing Platform.1
Individual-level genetic data describes the allele (e.g., A, C, G, T) found at each position of interest in the genome for each person in a group. While useful and highly detailed, this way of storing data leaves open the possibility of an individual’s genetic information being isolated and analyzed by bad actors. Individuals are always anonymized before being saved in a data format like this, but this still poses an unnecessary risk. In summary statistics data, all individuals in the study are grouped and correlations are carried out (such as simple linear regression) to determine the effects of the alleles, in aggregate, on disease and prediction. Each variant’s correlation – sometimes called the effect size or beta – with the trait of interest, can be informative to trait prediction without requiring individual-level information. As an added bonus, such summary files are normally much smaller and easier to handle.
Drs. Jin and Bingxin have shown that these summary statistics can be used to develop effective PRS models, using pseudo-training that does not require individual level statistics. These new pseudo-training methods build on the PUMAS method developed by Drs. Zijie Zhao and Qiongshi Lu at the University of Wisconsin – Madison2. Using this PUMAS framework with special modifications, PennPRS additionally combines multiple PRS models into an ensemble model. When evaluating the effectiveness of the ensemble PennPRS compared to individual-level models, Jin et. al. found that the pseudo-training methods performed comparably, with similar prediction R2 values, which measure overall model predictability. Importantly, in cases of insufficient individual data for model tuning, “pseudo PRS training notably outperforms traditional PRS training methods that rely on individual-level tuning data.”1
The authors acknowledge that some scientists may already have access to high computing resources and could be hesitant to use web-based tools. PennPRS has an offline version available for download to account for this, which is also a more convenient choice for large-scale PRS training compared to online training, but the authors note that using the resource in this way may be more time consuming and less environmentally friendly. PennPRS is noted as a promising PRS option for smaller research groups with limited access to large computational resources.
Polygenic risk scores are an exciting part of genetics, allowing predictions of risk for various traits and diseases. However, roadblocks to widespread use can be computational, logistical, or cost-related, and largely due to complications introduced by individual-level GWAS data. Drs. Jin and Zhao have shown PennPRS as a promising method to gain the benefits of existing PRS models while using only GWAS summary statistics. Available as a web tool, PennPRS is accessible for training and prediction on summary-level data using cutting edge methods.
For a tutorial on how to use PennPRS, please see https://pennprs.gitbook.io/pennprs. For full details, please visit the pennprs.org website or manuscript, which has not yet been peer-reviewed, in medRxiv: https://doi.org/10.1101/2025.02.07.25321875.
References
- Jin, J., Li, B., et al. PennPRS: a centralized cloud computing platform for efficient polygenic risk score training in precision medicine. MedRxiv. (2025) https://doi.org/10.1101/2025.02.07.25321875.
- Zhao, Z., Yi, Y., Song, J. et al. PUMAS: fine-tuning polygenic risk scores with GWAS summary statistics. Genome Biol 22, 257 (2021). https://doi.org/10.1186/s13059-021-02479-9