By Ulli Hain, Lumina Corps.
As a statistical geneticist, Dr. Jung-Ying Tzeng, Professor of Statistics at North Carolina State University and an affiliate faculty member of the Penn Neurodegeneration Genomics Center (PNGC) faculty, enjoys tackling big challenges in omics.
“The work is motivated by what’s encountered in real life. You look at the needs from the biological side and come up with solutions,” says Dr. Tzeng. “I appreciate that the field of genetics moves rapidly so there are always new challenges.”
Nowhere are these challenges truer than in Alzheimer’s disease (AD), where about half of the heritability in the late onset form remains unaccounted for. One area ripe for exploration is copy number variations (CNVs), which are segments of the genome that are deleted or duplicated. Ranging from ten base pairs to megabases, they can change the “dosage” of genes and either protect against or increase the risk of disease.
CNV analysis is complex, as both the length of the CNV and the number of copies matter. Because CNVs span multiple base pairs and may only partially overlap between people, there is no natural definition for a CNV locus. Yet, conventional kernel association methods, useful tools for aggregate analysis for rare CNVs, require one to arbitrarily define the CNV locus, which can lead to inconsistencies between studies.
“Each of us has CNVs at different beginning and end positions so they’re not well aligned across individuals,” said Dr. Tzeng. “We should respect that that’s what nature looks like and consider it in the analysis.”
To address this issue, Dr. Tzeng’s group developed a curve-based approach called CONCUR that eliminates the CNV locus definition requirement. Instead, an individual’s CNV information is modeled as continuous events over a genomic region. It is depicted in a “CNV profile curve,” where a straight line represents the baseline of two copy numbers, and a duplication is visualized as a hump and a deletion as a trough.
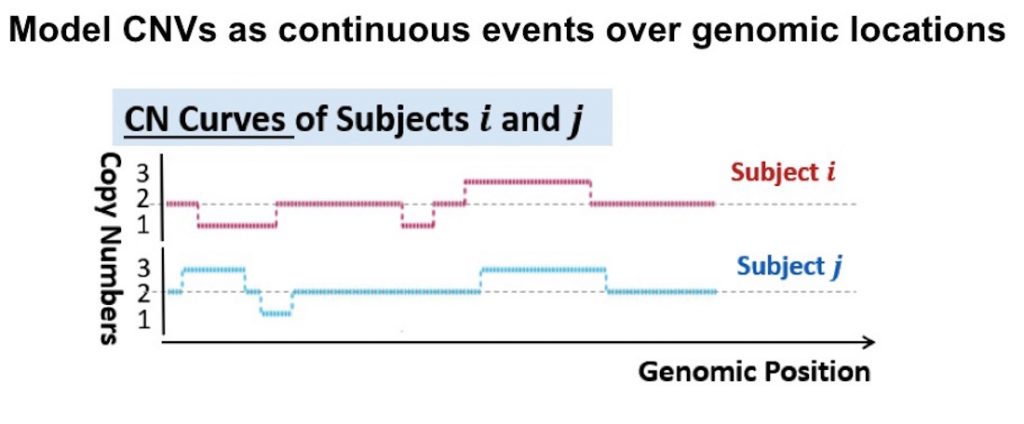
These curves are superimposed between individuals to quantify the CNV similarities via the common area under the curve, which takes into account both CNV length and number of copies. A score-based variance component test then determines the association between the CNV profiles and the phenotype—in this case, AD status.
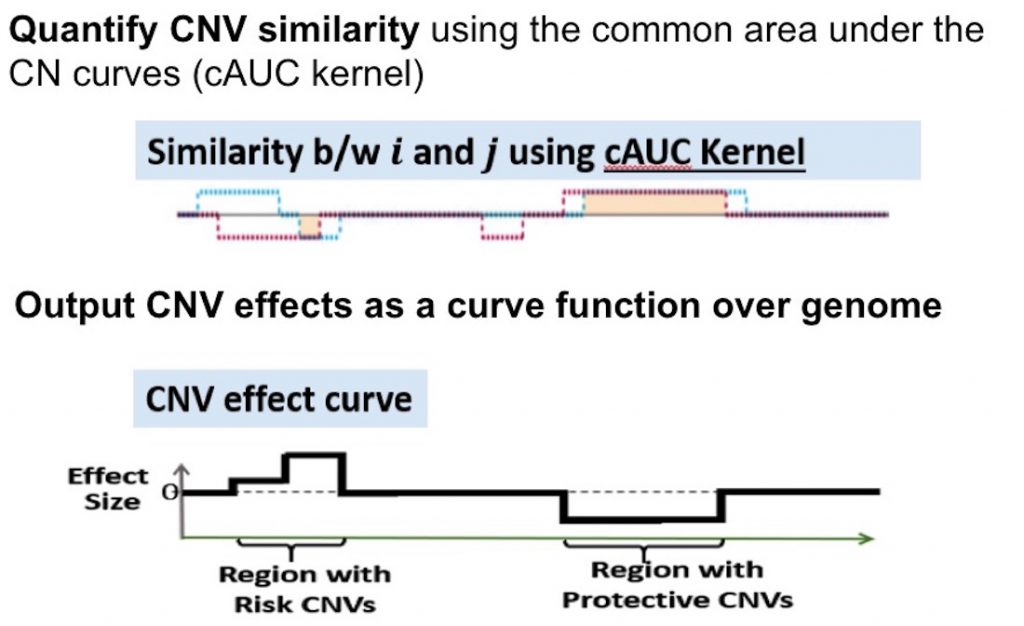
The curve-based approach has implications for all CNV analyses, not just those related to AD. In a series of test analyses published in 2020, the curve-based CONCUR method outperformed other kernel-based approaches. CONCUR is freely available as an R-package on CRAN.
Dr. Tzeng is working with PNGC faculty member Dr. Wan-Ping Lee, Research Assistant Professor of Pathology and Laboratory Medicine at the University of Pennsylvania, to apply this method to the recently available whole genome sequencing (WGS) data from the Alzheimer’s Disease Sequencing Project (ADSP). Although most Alzheimer’s CNV studies used data from genotyping arrays, WGS allows for a more unbiased approach to find more types of CNVs, such as small and rare ones, that aren’t possible with arrays.
Last year, the team was awarded an NIH grant to identify and study new CNVs associated with AD. New CNVs will provide insights about disease mechanisms and new therapeutic targets for AD.
In addition to assessing the collective effect of rare CNVs on AD, Drs. Tzeng and Lee plan to look at the joint effect of these CNVs and co-localized or nearby single-nucleotide variants (SNVs) and short insertions/deletions (INDELs).
“It is exciting that ADSP generates such large-scale WGS data for a single human disease,” said Dr. Lee. “It challenges us scientists to consider all kinds of genomic variants, including SNVs, INDELs, and CNVs together and to combine that information with other omics data to decipher Alzheimer’s.”
Beyond these analyses, Dr. Tzeng works with Dr. Lee and PNGC Co-Director Dr. Li-San Wang on improving methods to determine gene-environment interactions in AD using biobank data or other large datasets. Since joining the team in 2019, Dr. Tzeng has found the collegial environment of the PNGC refreshing.
“It is great to develop methods in conjunction with a team, where at every step, we can discuss what the project needs while also ensuring the approaches will be accessible to a majority of users,” said Dr. Tzeng. “In the past, I would have made my best educated guess, but now I can discuss with Wan-Ping and Li-San. There are among those rare people with whom one feels comfortable asking questions, pointing out different thoughts on an issue, and finding a way to move forward.”