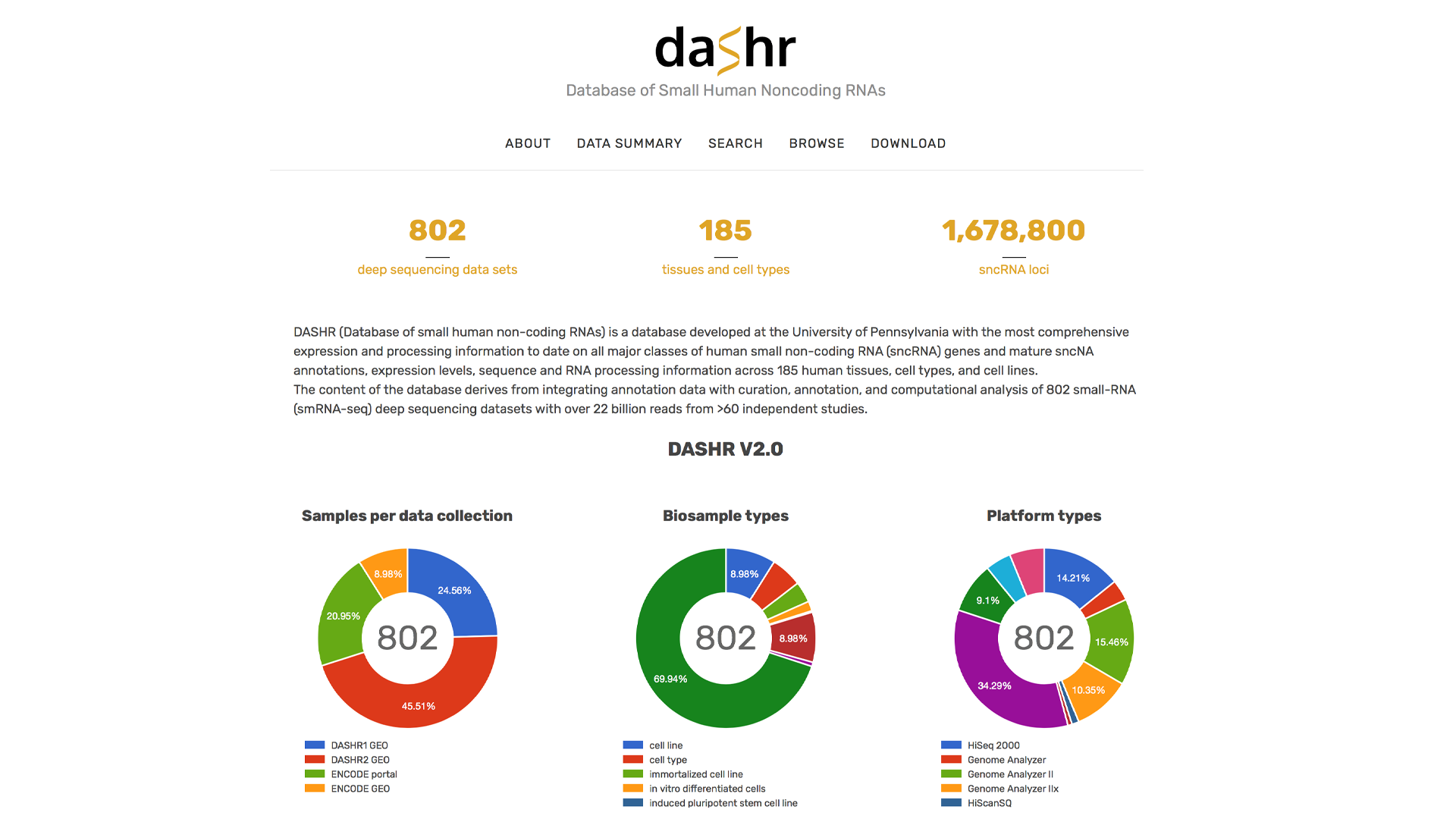
PNGC is pleased to announce the release of the DASHR v2.0 database of human small non-coding RNAs (sncRNAs). DASHR v2.0 is freely available at https://lisanwanglab.org/DASHRv2. This is an extremely useful resource and reference for the community studying non-coding RNAs and their diverse roles in often tissue-specific cellular processes related to human diseases.
DASHR (Database of small human non-coding RNAs) v2.0 provides a complete catalog of annotation, expression, processing, conservation, tissue-specificity and other biological features for all human sncRNA genes, full sncRNA transcripts and mature products derived from all major small RNA classes, including miRNAs, piRNAs, sc-, sn-, sno- RNAs, tRNAs, tRFs.
Some of the DASHR v2.0 highlights include:
1) Integration of the largest collection of >180 tissues and cell types from ENCODE and GEO/SRA across multiple RNA-seq protocols for both GRCh38/hg38 and GRCh37/hg19;
2) The first database to profile both known sncRNA loci and novel, previously un-annotated sncRNA loci identified by genome-wide unsupervised segmentation;
3) Integration of >3,200,000 annotations for non-small RNA genes and other genomic features (e.g., long-noncoding RNAs, promoters);
4) Introduction of an enhanced user interface, interactive experiment-by-locus table views for easy sorting and filtering of small RNA loci data;
5) Access to genome wide expression tracks and called small RNA peaks freely downloadable and viewable in the UCSC Genome Browser.
DASHR 2.0 has been published in Bioinformatics.
Kuksa, Pavel P., Amlie-Wolf, A., Katanic, Z., Valladares, O., Wang, L., Leung, Y. DASHR v2.0: database of small human noncoding RNAs in human tissues and cell types. Bioinformatics, 2018. https://doi.org/10.1093/bioinformatics/bty709